All issues
Author:Wen-Li Lee, Chinnapan Thanarut, Chih-Cheng Hsu, Guo-Chi Lee, Fen-Lang Chiang, and Chin-Chih Chen*
Abstract:
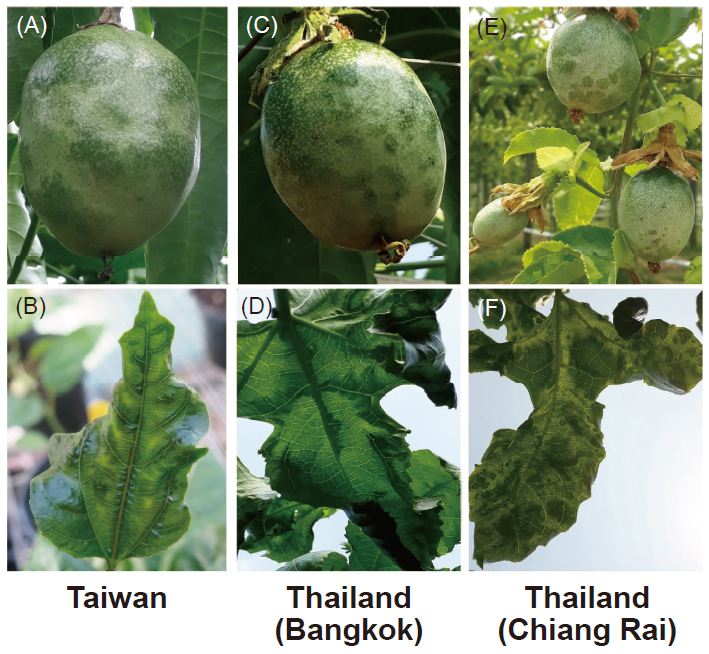
In this study, a polyclonal antibody against Telosma mosaic virus (Ab-TeMV) was prepared from the bacteria expressed TeMV coat protein gene. Ab-TeMV had a broad-spectrum detection efficiency for the potyviruses of TeMV, East Asian passiflora virus (EAPV), Cowpea aphid borne mosaic virus (CABMV), Bean yellow mosaic virus (BYMV) and Soybean mosaic virus (SMV). The full-length genomic nucleotide sequences of TeMV-DC6, an isolate infecting passiflora in Taiwan, was determined (GenBank accession number MN316594). The incidence of TeMV on passiflora in fields of Taiwan during 2017– 2019 was 32.1 to 35.8% by serological and molecular detection reagents of TeMV prepared in this study. Moreover, only TeMV was positively detected from those 17 virus-infected passiflora samples (showed symptoms of leaf mosaic and woodiness fruit) collected from Thailand. The CP amino acid sequences of those TeMV isolates collected from Taiwan and Thailand’s passifloa fields shared higer than 90% identities with that of the known TeMV (AM409188) in GenBank. However, the phylogenetic analysis indicated that TeMV isolates of passiflora from different geographical locations are different in phylogenetic evolution, because 9 Taiwan isolates are closely related and separated from those 17 Thailand isolates. Three sets of primer pairs were designed based on the aligned nucleotide sequences of Taiwan and Thailand TeMVs, and used in reverse transcription-polymerase chain reaction (RT-PCR). Among the 67 Taiwan isolates and 17 Thailand isolates, the Taiwan primer pair (TwTe650u/TwTe650d) can amplify the expected amplicons from all of the tested Taiwan isolates and 9 out of the 17 Thailand isolates. The Thailand primer pair (ThaTe430u/ThaTe430d) only detected 12 out of the 17 Thailand isolates and could not detect all of the Taiwan isolates. The Taiwan-Thailand primer pair (TwTha840u/TwTha840d) could detect all of the tested isolates. In this report, the incidences of TeMV in Taiwan and Thailand were clarified and the full-length genomic sequence of Taiwan isolate DC6 was first completed. The phylogenetic differences between Taiwan and Thailand TeMV isolates were demonstrated, and the primer pairs were developed for the wide-spectrum detection of those passiflora TeMV isolates in Taiwan and Thailand to improve the detection efficacy of TeMV.
Key words:Passiflora sp., Telosma mosaic virus (TeMV), Polyclonal antibody, Full-length sequence, Broad-spectrum detection by RT-PCR
Download:![]() PDF Links
PDF Links
- 1. Using Digital Soil Mapping to Predict Soil Organic Carbon Stocks in Zhuoshui River Basin
- 2. Taxonomic Review of the Genus Asiophrida Medvedev, 1999 in Taiwan (Insecta: Coleoptera: Chrysomelidae: Galerucinae: Alticini), with Notes on Biology
- 3. Development of a Technique for Forecasting (or Pre-Detection) Anthracnose Disease Incidences of Green Mature Bagging Mango Fruits
 Submit your manuscript
Submit your manuscript
 Guide for authors
Guide for authors