All issues
Author:Cheng-Ping Kuan, Ying-Chuan Chiang, Chung-Jen Hsiao, and Ching-Shan Tseng*
Abstract:
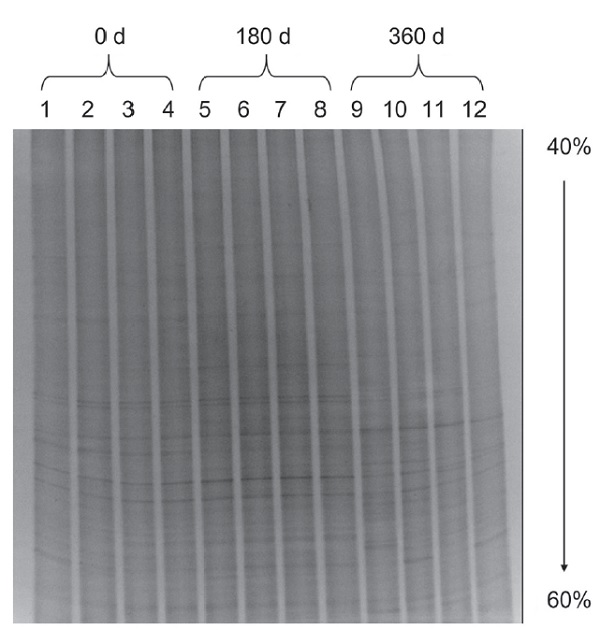
To understand the impact of buried rice straw residues of transgenic rice (AAN), which overexpresses phytase enzymes on the soil microbial community, a year-long monitoring experiment was conducted to track changes in the soil microbial composition. The results revealed that after 180 d of burial, the total bacterial count in the non-transgenic control variety ‘TNG 67’ (Oryza sativa L. ssp. japonica cv. Tainung 67; ‘TNG 67’) was significantly higher than that of the AAN transgenic line, and a declining trend was found along with time for both varieties. The total fungal count in the soil exhibited greater variability along with burial time. Although the AAN transgenic line generally had higher fungal counts, the difference between the two was not statistically significant after 120 d of burial. Notably, the total fungal count in the AAN transgenic line decreased significantly at 360 d. Regarding actinomycetes in the soil, the total count gradually increased over time in the ‘TNG 67’ control rice. However, an obvious increase was found in the AAN transgenic line after 120 d decomposition. Nevertheless, counts of both varieties began to decline after 300 d, and by 360 d, there was no significant difference between the two. Analysis using denaturing gradient gel electrophoresis (DGGE) showed an increased abundance of microbial communities near the AAN transgenic line. However, the bacterial communities derived from the AAN transgenic line and ‘TNG 67’ rice were found in the same cluster with high similarity at 180 d. The same result was found at 360 d in which the AAN transgenic line and ‘TNG 67’ bacterial communities exhibited similarity within the same cluster. Integrating the results from plate counting and DGGE analysis suggests that during the early stages of straw burial, there were notable differences in microbial count and bacterial diversity between the AAN transgenic line and the ‘TNG 67’ control. However, these differences gradually diminished or converged after 360 d of burial.
Key words:Rice, Transgenic crop, Soil bacterial community, Rice straw
Download:![]() PDF Links
PDF Links
- 1. Using Digital Soil Mapping to Predict Soil Organic Carbon Stocks in Zhuoshui River Basin
- 2. Development of a Technique for Forecasting (or Pre-Detection) Anthracnose Disease Incidences of Green Mature Bagging Mango Fruits
- 3. Taxonomic Review of the Genus Asiophrida Medvedev, 1999 in Taiwan (Insecta: Coleoptera: Chrysomelidae: Galerucinae: Alticini), with Notes on Biology
 Submit your manuscript
Submit your manuscript
 Guide for authors
Guide for authors