All issues
Author:Yann-Rong Lin, Hsiang-Ting Chien, Han-Shiuan Chin, Hung-Ying Lin, Hsing-Mu Yen, Woei-Shyuan Jwo, Dong-Hong Wu, Ming-Hsing Lai, and Charng-Pei Li*
Abstract:
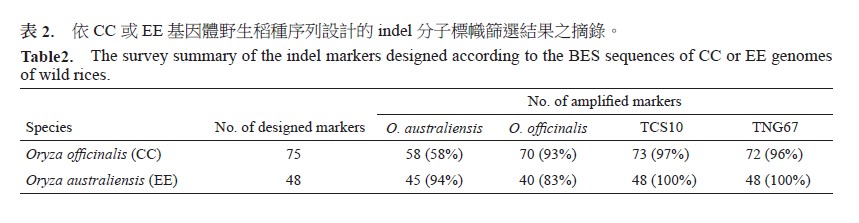
Wild rice has high genetic diversity and high tolerance to extreme environments. If genomes of wild rice can be applied into modern rice breeding programs by marker-assisted selection, newly bred cultivars can endure mercurial climate and meet sharply increased demands of food in the world. The goal for this research is to introgress genes/chromosome segments of Oryza officinalis with CC genome and Oryza australiensis with EE genome into Oryza sativa ssp. indica cv. ‘TCS10’ and ssp. japonica cv. ‘TNG67’ with AA genome, respectively. Currently, polymorphic markers were surveyed and developed. A total of 249 SSR (simple sequences repeat) had been applied but only 100 (40%) and 67 (27%) of markers could be amplified successfully in O. officinalis and O. australiensis, respectively. Among polymorphic SSR markers, 72 (72%) and 71 (71%) of markers were polymorphic between O. officinalis vs. ‘TCS10’ and ‘TNG67’, respectively; 57 (76%) and 53 (79%) of markers were polymorphic between O. australiensis vs. ‘TCS10’ and O. australiensis vs. ‘TNG67’, respectively. To effectively obtain polymorphic markers, the BES (BAC end sequence) of wild and the genome sequence of ssp. japonica cv. ‘Nipponbare’ were aligned to search for indel markers flanking with conserved sequences. A total of 123 markers had been developed, and 109 (89%) and 103 (84%) markers could be amplified successfully in O. officinalis and O. australiensis, respectively. Among which, 79 (72%) and 82 (75%) of markers were polymorphic between O. officinalis vs. ‘TCS10’ and ‘TNG67’, respectively; 64 (62%) and 65 (63%) of markers were polymorphic between O. australiensis vs. ‘TCS10’ and O. australiensis vs. ‘TNG67’, respectively. From genotypes of BC1F1 and BC2F1 populations derived from O. officinalis × ‘TNG67’, the ratio of heterozygotes was 86.7% (BC1F1) and abruptly decreased to 18.1% (BC2F1). It indicated that the chromosome segments of wild rice were excluded rapidly in backcrossed progenies, implying that it is very difficult to develop a complete chromosome segment substitute line (CSSL) population, which carried the chromosome segment from different genome of wild species into O. sativa background.
Key words:Molecular marker, Transferability, Rice, Wild rice
Download:![]() PDF Links
PDF Links
- 1. Development of Tractor-Mounted Seedling Transplanter for Sweet Potato
- 2. Synergistic Effect of Additional Gas on the Toxicity of Phosphine to Sitophilus oryzae and Sitophilus zeamais (Coleoptera: Dryophthoridae)
- 3. Effects of Temperature and Solar Radiation on Growth Traits and Plant Elements in Purple Leafy Sweet Potato
 Submit your manuscript
Submit your manuscript
 Guide for authors
Guide for authors