All issues
Author:Ching-Yi Lin, Chiao-Wen Huang, Hong-Ren Yang, Su-Yu Lai, and Hui-Fang Ni*
Abstract:
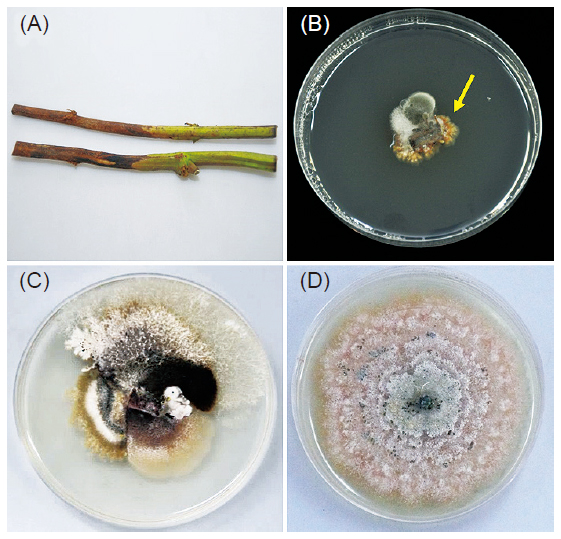
Foot rot caused by Phomopsis destruens (Harter) Boerema is a major concern for sweet potato production in Taiwan. The conventional methods for the identification of P. destruens are based on culture isolation and morphological analyses, but they are time-consuming and often inconclusive. To assist in a rapid and specific detection of the infection, a polymerase chain reaction (PCR) assay was developed to detect the pathogen from DNA extracted from fungi and plant tissues, respectively. Internal transcribed spacer (ITS) regions of the ribosomal DNA (rDNA) from P. destruens isolates were used for the specific primers design. The amplification of expected 298 bp PCR products was obtained in all P. destruens isolates, but not for other fungal isolates from sweet potato storage roots including Lasiodiplodia theobrome (B2225), Phomopsis sp. (Ph735), Rhizopus sp., Athelia rolfsii (Sc-3) and Fusarium sp. (Fu245). The PCR assay with the primers could detect 1 ng of DNA template. To enhance the sensitivity of detection, a nested PCR was performed and the detection limit was raised up to 10 pg. In addition, the primers proved efficient in detection of the pathogen in stems and storage roots of infected sweet potato. Positive detection was observed in 93% of the naturally infected stems of sweet potato, a rate which constituted a significant improvement in the identification of P. destruens in sweet potato as compared to conventional methods. These results indicated that the new PCR assay provided a rapid, specific and reliable diagnosis tool for the detection of sweet potato foot rot pathogen.
Key words:Ipomoea batatas, Phomopsis destruens, Foot rot, PCR assays
Download:![]() PDF Links
PDF Links
- 1. Development of Tractor-Mounted Seedling Transplanter for Sweet Potato
- 2. Synergistic Effect of Additional Gas on the Toxicity of Phosphine to Sitophilus oryzae and Sitophilus zeamais (Coleoptera: Dryophthoridae)
- 3. Effects of Temperature and Solar Radiation on Growth Traits and Plant Elements in Purple Leafy Sweet Potato
 Submit your manuscript
Submit your manuscript
 Guide for authors
Guide for authors