All issues
Author:Mei-Ju Lin, Yi-Ting Lin, Chin-Hui Tsai, and Ting-Chin Deng*
Abstract:
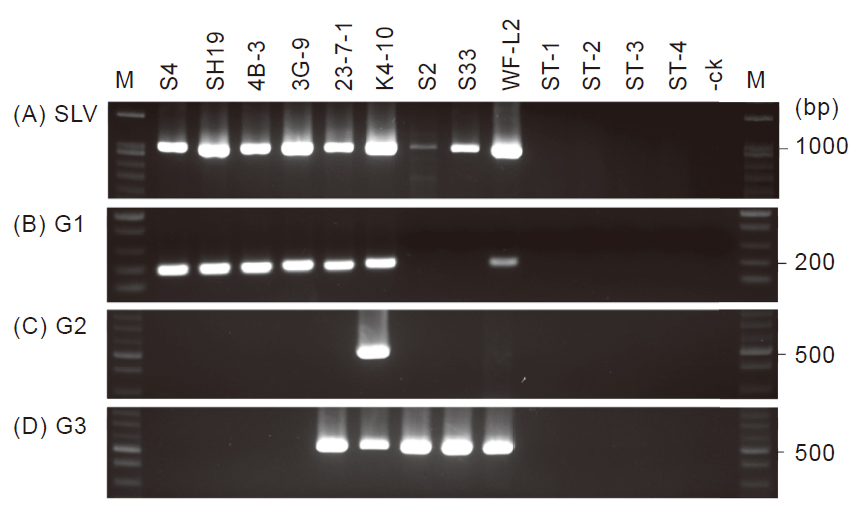
The coat protein (CP) gene nucleotide sequences of species Shallot latent virus (SLV) collected from National Center for Biotechnology Information (NCBI)-GenBank database were tested for phylogenetic analysis. All the current SLV isolates tested were clustered into three groups: G1, G2 and G3. In 2015–2016, a total of 10 SLV-infected Allium materials were collected from a survey of SLV occurred in Taiwan by detecting the virus with enzyme-linked immunosorbent assay (ELISA) and reverse transcription-polymerase chain reaction (RT-PCR). The CP genes of each isolates were cloned and sequenced, among 10 isolates tested, 6 isolates were clustered into G1 with 894 bp nucleotides. The others belong to G3 with 885 bp nucleotides of CP gene. Based on divergence of CP nucleotide sequences, the group specific primer pairs were designed to differentiate the groups. Following by a RT-PCR, the nested-polymerase chain reaction (nested-PCR) was performed, and the amplified DNA was 187 bp, 548 bp and 540 bp for G1, G2 and G3, respectively. Accordingly, 134 SLV-infected samples were collected and detected by RT-PCR and nested-PCR as described above. There were 51 out of 52 garlic bulbs and 24 out of 25 garlic plants were identified as G1. Moreover, a total of 11 samples from Chinese leeks were defined as G3, and 9 samples out of them were complex with G1 and G3. The other 34 samples tested from green onions were consisted of 5 belonged G1, 19 belonged G3 and 15 were complex with G1 and G3. In this study, no SLV isolates belonged G2 were found from Allium in Taiwan.
Key words:Shallot latent virus, Allium species, Phylogenetic analysis, Nested-polymerase chain reaction
Download:![]() PDF Links
PDF Links
- 1. Development of Tractor-Mounted Seedling Transplanter for Sweet Potato
- 2. Synergistic Effect of Additional Gas on the Toxicity of Phosphine to Sitophilus oryzae and Sitophilus zeamais (Coleoptera: Dryophthoridae)
- 3. Effects of Temperature and Solar Radiation on Growth Traits and Plant Elements in Purple Leafy Sweet Potato
 Submit your manuscript
Submit your manuscript
 Guide for authors
Guide for authors