All issues
Author:Jau-Yeuh Wang, Chia-Wei Song, Min-Huai Liou and Shyi-Kuan Ou*
Abstract:
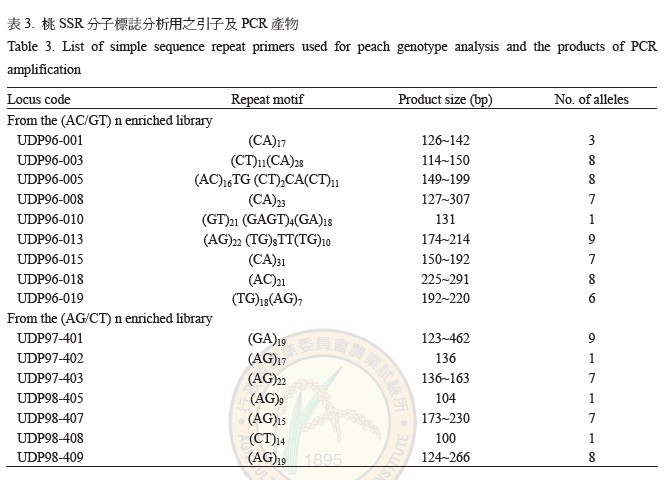
The typical fruit characteristics of 34 selected peach cultivars and the hybrid seedlings were selected for genetic similarity analysis and identification by microsatellite DNA markers (including SSR: simple sequence repeat; and ISSR: inter simple sequence repeat). The selected typical fruit characteristics included melting, non-melting and stony hard types, sour versus bitter, sweet versus very sweet and with versus without pubescence. Amplification of SSR locus was screened from 17 primer pairs and 12 polymorphic SSR markers had been selected which had 3 to 9 alleles each. One of the SSR markers UPD-96-005 could be used to distinguish the domestic cultivar of ‘Ying Ge Tao’ and ‘Ba Yue Tao’. Furthermore, combining all of the 87 SSR markers and assessing the genetic similarity by cluster analysis of UPGMA (un-weighted pair-group mean arithmetic). It revealed that 34 genotypes could be divided into two main groups: the first group contained 16 genotypes including ‘Ying Ge Tao’ and its hybrid progenies; the second group was composed by 18 genotypes including ‘Premier’ and its hybrid progenies. From the dendrogram of UPGMA analysis showed the progenies would be grouping with parent and accompanied with the level of TTS (total soluble solid) and acidity separately. The results reveal that microsatellite DNA markers are useful in cultivars identification of peach and allow to early selection of the progenies with superior fruit characteristics.
Key words:Peach, Microsatellite DNA, Genetic similarity, Fruit characteristics
Download:![]() PDF Links
PDF Links
- 1. Development of Tractor-Mounted Seedling Transplanter for Sweet Potato
- 2. Synergistic Effect of Additional Gas on the Toxicity of Phosphine to Sitophilus oryzae and Sitophilus zeamais (Coleoptera: Dryophthoridae)
- 3. Effects of Temperature and Solar Radiation on Growth Traits and Plant Elements in Purple Leafy Sweet Potato
 Submit your manuscript
Submit your manuscript
 Guide for authors
Guide for authors