All issues
Author:Meng-Li Wei, Yi-Han Chung, Chun-Tang Lu, and Hsiu-Ying Lu*
Abstract:
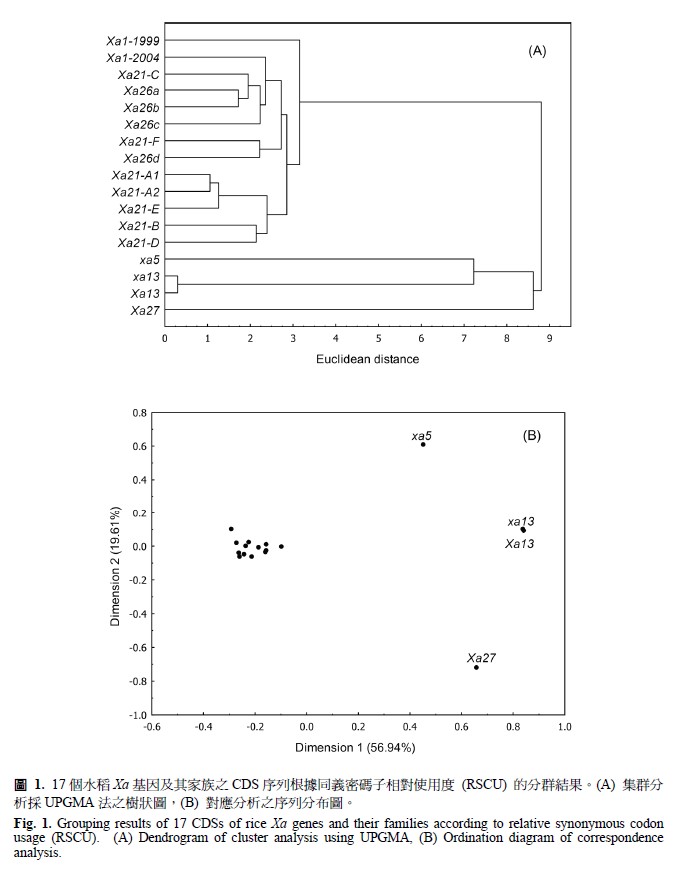
Analysis of codon usage is important in understanding the molecular mechanism of rice genes with resistance to bacterial blight caused by Xanthomonas oryzae pv. oryzae. A total of 17 coding domain sequences (CDSs) of the completely sequenced genes, i.e., Xa1, xa5, xa13, Xa13, Xa21, Xa26, and Xa27 were obtained through the access from public database of NCBI (National Center for Biotechnology Information) and they were used to analyze the condon usage bias. The use of correspondence analysis and cluster analysis of relative synonymous codon usage (RSCU) clearly separated the CDSs of Xa genes into four distinct groups. The relative frequency of synonymous codon (RFSC) was further used to indentify the high-frequency codons for each gene group. The results showed that Xa1, Xa21, and Xa26 with leucine-rich repeat (LRR) domain protein structure and CDS length more than 1000 bp were found no codon usages in most amino acids, and preferred A- or U-ending codons in a few amino acids; whereas xa5, xa13, Xa13, and Xa27 with non-LRR domain protein structure and shorter CDSs preferred C- or G-ending codons in almost all amino acids. The protein sequences of Xa1, Xa21, Xa26, and Xa27 has been known to have abundant leucine (Leu). The Xa1, Xa21, and Xa26 showed no significant codon bias in their Leu, that is they could choose any synonymous codon for Leu when translating to protein; whereas, Xa27 exhibited CUC as the most preferred codon in Leu. It indicated that codon usage bias varied among rice genes with resistance to bacterial blight.
Key words:Xa genes, Relative synonymous codon usage, Relative frequency of synonymous codon, Cluster analysis, Correspondence analysis
Download:![]() PDF Links
PDF Links
- 1. Development of Tractor-Mounted Seedling Transplanter for Sweet Potato
- 2. Synergistic Effect of Additional Gas on the Toxicity of Phosphine to Sitophilus oryzae and Sitophilus zeamais (Coleoptera: Dryophthoridae)
- 3. Effects of Temperature and Solar Radiation on Growth Traits and Plant Elements in Purple Leafy Sweet Potato
 Submit your manuscript
Submit your manuscript
 Guide for authors
Guide for authors